This tutorial will show you how to implement a K-Nearest Neighbors algorithm for classification in Python. We are going to visualize a data set, find the best parameters to use, train a model and evaluate the results. K-Nearest Neighbors is a simple algorithm to understand and it usually gets very high accuracy scores.
K-Nearest Neighbors for classification gathers the K closest observations and returns the most common class among these. A K-Nearest Neighbors model needs training data for predictions and this means that the model will be large in size if the training data set is large. This algorithm is not good at making predictions on input values that are outside of the training data set, you can continuously add more training data to keep the model up to date. A K-Nearest Neighbors model is complex as it fits the training data quite well but might be bad a generalization on new and unseen data.
Data set and libraries
I am going to use the Iris flower data set (download it) in this tutorial, a small toy data set that is used for learning. The Iris flower data set consists of 150 flowers, each flower has four input values and one output value. I am also using the following libraries: pandas, joblib, numpy, matplotlib and scikit-learn. You need to install these libraries with pip.
Python module
I have included all code in one file, a project normally consists of many files (modules). You can create namespaces by placing files in folders and import a file by its namespaces plus its file name. A file named common.py in a annytab/learn folder is imported as import annytab.learn.common. I am going to explain more about the code in sections below.
# Import libraries
import pandas
import joblib
import numpy as np
import matplotlib.pyplot as plt
import sklearn.model_selection
import sklearn.neighbors
import sklearn.metrics
import sklearn.pipeline
# Visualize dataset
def visualize_dataset(ds):
# Print first 5 rows in dataset
print('--- First 5 rows ---\n')
print(ds.head())
# Print the shape
print('\n--- Shape of dataset ---\n')
print(ds.shape)
# Print class distribution
print('\n--- Class distribution ---\n')
print(ds.groupby('species').size())
# Box plots
plt.figure(figsize = (12, 8))
ds.boxplot()
#plt.show()
plt.savefig('plots\\iris-boxplots.png')
plt.close()
# Scatter plots (4 subplots in 1 figure)
figure = plt.figure(figsize = (12, 8))
grouped_dataset = ds.groupby('species')
values = ['sepal_length', 'sepal_width', 'petal_length', 'petal_width']
for i, value in enumerate(values):
plt.subplot(2, 2, i + 1) # 2 rows and 2 columns
for name, group in grouped_dataset:
plt.scatter(group.index, ds[value][group.index], label=name)
plt.ylabel(value)
plt.xlabel('index')
plt.legend()
#plt.show()
plt.savefig('plots\\iris-scatterplots.png')
# Train and evaluate
def train_and_evaluate(X, Y):
# Create a model
model = sklearn.neighbors.KNeighborsClassifier(n_neighbors=10, weights='distance', algorithm='auto', leaf_size=10, p=2, metric='minkowski', metric_params=None, n_jobs=1)
# Train the model on the whole dataset
model.fit(X, Y)
# Save the model (Make sure that the folder exists)
joblib.dump(model, 'models\\knn.jbl')
# Evaluate on training data
print('\n-- Training data --\n')
predictions = model.predict(X)
accuracy = sklearn.metrics.accuracy_score(Y, predictions)
print('Accuracy: {0:.2f}'.format(accuracy * 100.0))
print('Classification Report:')
print(sklearn.metrics.classification_report(Y, predictions))
print('Confusion Matrix:')
print(sklearn.metrics.confusion_matrix(Y, predictions))
print('')
# Evaluate with 10-fold CV
print('\n-- 10-fold CV --\n')
predictions = sklearn.model_selection.cross_val_predict(model, X, Y, cv=10)
accuracy = sklearn.metrics.accuracy_score(Y, predictions)
print('Accuracy: {0:.2f}'.format(accuracy * 100.0))
print('Classification Report:')
print(sklearn.metrics.classification_report(Y, predictions))
print('Confusion Matrix:')
print(sklearn.metrics.confusion_matrix(Y, predictions))
# Perform a grid search to find the best parameters
def grid_search(X, Y):
# Create a pipeline
clf_pipeline = sklearn.pipeline.Pipeline([
('m', sklearn.neighbors.KNeighborsClassifier(metric='minkowski', metric_params=None, n_jobs=1))
])
# Set parameters (name in pipeline + name of parameter)
parameters = {
'm__n_neighbors': (1, 2, 3, 4, 5, 6, 7, 8, 9, 10),
'm__weights': ('uniform', 'distance'),
'm__algorithm': ('auto', 'ball_tree', 'kd_tree', 'brute'),
'm__leaf_size': (10, 20, 30),
'm__p': (1, 2)
}
# Create a grid search classifier
gs_classifier = sklearn.model_selection.GridSearchCV(clf_pipeline, parameters, cv=5, iid=False, n_jobs=2, scoring='accuracy', verbose=1)
# Start a search (Warning: can take a long time if the whole dataset is used)
gs_classifier = gs_classifier.fit(X, Y)
# Print results
print('---- Results ----')
print('Best score: ' + str(gs_classifier.best_score_))
for name in sorted(parameters.keys()):
print('{0}: {1}'.format(name, gs_classifier.best_params_[name]))
# Predict and evaluate on unseen data
def predict_and_evaluate(X, Y):
# Load the model
model = joblib.load('models\\knn.jbl')
# Make predictions
predictions = model.predict(X)
# Print results
print('\n---- Results ----')
for i in range(len(predictions)):
print('Input: {0}, Predicted: {1}, Actual: {2}'.format(X[i], predictions[i], Y[i]))
accuracy = sklearn.metrics.accuracy_score(Y, predictions)
print('\nAccuracy: {0:.2f}'.format(accuracy * 100.0))
print('\nClassification Report:')
print(sklearn.metrics.classification_report(Y, predictions))
print('Confusion Matrix:')
print(sklearn.metrics.confusion_matrix(Y, predictions))
# The main entry point for this module
def main():
# Load dataset (includes header values)
dataset = pandas.read_csv('files\\iris.csv')
# Visualize dataset
visualize_dataset(dataset)
# Slice dataset in values and targets (2D-array)
X = dataset.values[:,0:4]
Y = dataset.values[:,4]
# Split dataset in train and test (use random state to get the same split every time, and stratify to keep balance)
X_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X, Y, test_size=0.2, random_state=1, stratify=Y)
# Make sure that data still is balanced
print('\n--- Class balance ---\n')
print(np.unique(Y_train, return_counts=True))
print(np.unique(Y_test, return_counts=True))
# Perform a grid search
#grid_search(X_train, Y_train)
# Train and evaluate
#train_and_evaluate(X_train, Y_train)
# Predict on testset
predict_and_evaluate(X_test, Y_test)
# Tell python to run main method
if __name__ == "__main__": main()Load and visualize the data set
The data set is loaded with pandas by using an relative path to the root of the project, use an absolute path if your files is stored outside of the project. We want to visualize the data set to make sure that it is balanced and we want to learn more about the data. It is important to have a balanced data set when performing classification, every class will be trained equally many times with a balanced training set. We can plot a data set to find patterns, remove outliers and decide on the most suitable algorithms to use.
# Load dataset (includes header values)
dataset = pandas.read_csv('files\\iris.csv')
# Visualize dataset
visualize_dataset(dataset)--- First 5 rows ---
sepal_length sepal_width petal_length petal_width species
0 5.1 3.5 1.4 0.2 Iris-setosa
1 4.9 3.0 1.4 0.2 Iris-setosa
2 4.7 3.2 1.3 0.2 Iris-setosa
3 4.6 3.1 1.5 0.2 Iris-setosa
4 5.0 3.6 1.4 0.2 Iris-setosa
--- Shape of dataset ---
(150, 5)
--- Class distribution ---
species
Iris-setosa 50
Iris-versicolor 50
Iris-virginica 50
dtype: int64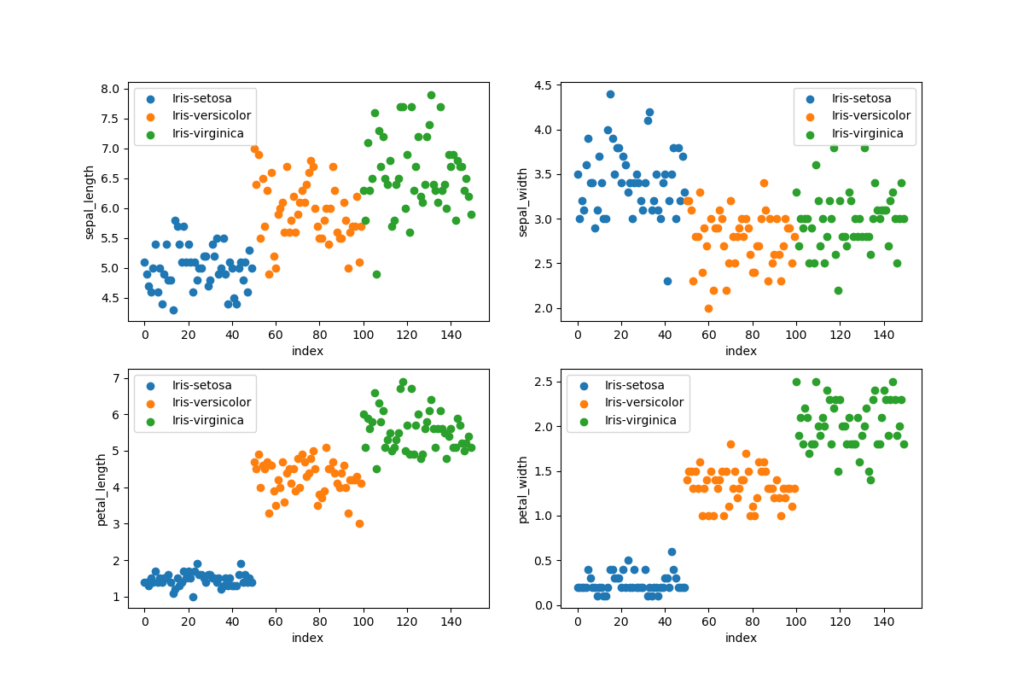
Split data set
I first need to slice values in the data set to get input values (X) and output values (Y), the first 4 columns is input values and the last column is the target value. I split the data set in a training set and a test set, 80 % is for training and 20 % for test. I want to make sure that data sets still are balanced after this split and I therefore use a stratify parameter.
# Slice dataset in values and targets (2D-array)
X = dataset.values[:,0:4]
Y = dataset.values[:,4]
# Split dataset in train and test (use random state to get the same split every time, and stratify to keep balance)
X_train, X_test, Y_train, Y_test = sklearn.model_selection.train_test_split(X, Y, test_size=0.2, random_state=1, stratify=Y)
# Make sure that data still is balanced
print('\n--- Class balance ---\n')
print(np.unique(Y_train, return_counts=True))
print(np.unique(Y_test, return_counts=True))Baseline performance
Our data set has 150 flowers and 50 flowers in each class, our training set has the same balance. A random prediction will be correct in 33 % (50/150) of all cases and our model must have an accuracy that is better than 33 % to be useful.
GridSearch
I am doing a grid search to find the best parameters to use for training. A grid search can take a long time to perform on large data sets but it’s probably faster than a manual process. The ouput from this process is shown below and I am going to use these parameters when I train the model.
# Perform a grid search
grid_search(X, Y)
Fitting 5 folds for each of 480 candidates, totalling 2400 fits
[Parallel(n_jobs=2)]: Using backend LokyBackend with 2 concurrent workers.
[Parallel(n_jobs=2)]: Done 1030 tasks | elapsed: 2.6s
[Parallel(n_jobs=2)]: Done 2400 out of 2400 | elapsed: 4.4s finished
---- Results ----
Best score: 0.9866666666666667
m__algorithm: auto
m__leaf_size: 10
m__n_neighbors: 10
m__p: 2
m__weights: distanceTraining and evaluation
I am training the model by using the parameters from the grid search and saved the model to a file with joblib. Evaluation is made on the training set and with cross-validation. The cross-validation evaluation will give a hint on the generalization performance of the model. I had 100 % accuracy on training data and 96.67 % accuracy with 10-fold cross validation.
# Train and evaluate
train_and_evaluate(X_train, Y_train)
-- Training data --
Accuracy: 100.00
Classification Report:
precision recall f1-score support
Iris-setosa 1.00 1.00 1.00 40
Iris-versicolor 1.00 1.00 1.00 40
Iris-virginica 1.00 1.00 1.00 40
accuracy 1.00 120
macro avg 1.00 1.00 1.00 120
weighted avg 1.00 1.00 1.00 120
Confusion Matrix:
[[40 0 0]
[ 0 40 0]
[ 0 0 40]]
-- 10-fold CV --
Accuracy: 96.67
Classification Report:
precision recall f1-score support
Iris-setosa 1.00 1.00 1.00 40
Iris-versicolor 0.97 0.93 0.95 40
Iris-virginica 0.93 0.97 0.95 40
accuracy 0.97 120
macro avg 0.97 0.97 0.97 120
weighted avg 0.97 0.97 0.97 120
Confusion Matrix:
[[40 0 0]
[ 0 37 3]
[ 0 1 39]]Prediction and evaluation
The final step in this process is to make predictions and evaluate the performance on the test data set. I load the model, make predictions and print the results. The X variable is a 2D array, if you want to make a prediction on one flower you need to set the input like this: X = np.array([[7.3, 2.9, 6.3, 1.8]]).
# Predict on test set
predict_and_evaluate(X_test, Y_test)
---- Results ----
Input: [7.3 2.9 6.3 1.8], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [4.9 3.1 1.5 0.1], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [5.1 2.5 3.0 1.1], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [4.8 3.4 1.6 0.2], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [5.0 3.5 1.6 0.6], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [5.1 3.5 1.4 0.2], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [6.2 3.4 5.4 2.3], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [6.4 2.7 5.3 1.9], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [5.6 2.8 4.9 2.0], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [6.8 2.8 4.8 1.4], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [5.4 3.9 1.3 0.4], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [5.5 2.3 4.0 1.3], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [6.8 3.0 5.5 2.1], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [6.0 2.2 4.0 1.0], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [5.7 2.5 5.0 2.0], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [5.7 4.4 1.5 0.4], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [7.1 3.0 5.9 2.1], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [6.1 2.8 4.0 1.3], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [4.9 2.4 3.3 1.0], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [6.1 3.0 4.9 1.8], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [6.4 2.9 4.3 1.3], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [5.6 3.0 4.5 1.5], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [4.9 3.1 1.5 0.1], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [4.4 2.9 1.4 0.2], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [6.5 3.0 5.2 2.0], Predicted: Iris-virginica, Actual: Iris-virginica
Input: [4.9 2.5 4.5 1.7], Predicted: Iris-versicolor, Actual: Iris-virginica
Input: [5.4 3.9 1.7 0.4], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [4.8 3.0 1.4 0.1], Predicted: Iris-setosa, Actual: Iris-setosa
Input: [6.3 3.3 4.7 1.6], Predicted: Iris-versicolor, Actual: Iris-versicolor
Input: [6.5 2.8 4.6 1.5], Predicted: Iris-versicolor, Actual: Iris-versicolor
Accuracy: 96.67
Classification Report:
precision recall f1-score support
Iris-setosa 1.00 1.00 1.00 10
Iris-versicolor 0.91 1.00 0.95 10
Iris-virginica 1.00 0.90 0.95 10
accuracy 0.97 30
macro avg 0.97 0.97 0.97 30
weighted avg 0.97 0.97 0.97 30
Confusion Matrix:
[[10 0 0]
[ 0 10 0]
[ 0 1 9]]